Past Issues
Research Advances in Biosynthesis Mechanism of Ultra-High Adhesive Material - Holdfast
Qing Liu, Chao Zhang, Dong-gang Xu*
1Institute of Military Cognitive and Brain Sciences, Beijing, 100850, PR China
*Corresponding author: Dong-gang Xu, Institute of Military Cognitive and Brain Sciences, Beijing, Tel: +86-010-66931329; E-mail: [email protected].
Received: September 13, 2021
Published: October 27, 2021
ABSTRACT
As a unique secretary product of Caulobacter crescentus, holdfast is the most powerful natural viscous substance known so far, and its detailed synthetic mechanism is still not clear. The biosynthetic process of holdfast is complex, including the synthesis, transportation and secretion of monosaccharides and polysaccharides, as well as the various modifications in each above-mentioned step. Many genes related to the sugar metabolism, the synthesis and modification of proteins, the modification of polysaccharides, the transportation and the secretion of polymer are involved in this process. In addition, the production of holdfast is also affected by cell cycle and stress. This review focuses on the recent advances in the study on the physicochemical property, transportation, secretion and modification of holdfast, as well as its synthetic pathways. The complex associations of genes involved in holdfast biosynthesis are also summarized, which helps deepen the understanding of its biosynthetic mechanism, thus gives the guidance on the future research and development of holdfast.
KEYWORDS: Holdfast; Caulobacter crescentus; Biosynthesis; Deacetylation; Transportation and Secretion
INTRODUCTION
In the microbial world, holdfast, the natural substance with the strongest adhesion, presents in Caulobacter crescentus [1,2], and is not easily observed. As a natural adhesive material with high viscosity, high environmental adaptability, high biocompatibility and no chemical pollution, the application of holdfast is extensive, especially in précis ion instrument manufacturing and underwater engineering. Holdfast is an ultra-viscous polysaccharide, which is different from the natural adhesive substances commonly found in nature. For example, mussels secrete the foot filament proteins Mfp-3 and Mfp-5 interacted with the interface through their DOPA structure to produce viscosity [3-5]. Barnacles secrete the adhesive protein cp19k, which is a self- assembling fibrin resulted in the viscosity through the aggregation of its amyloid fibrils [6,7]. In general, polysaccharide substances such as chitosan, hyaluronic acid and dermatan sulfate are not highly viscous, which are usually developed into health care or medical products [8-11]. On the other hand, the discovery of holdfast breaks the perception that polysaccharides are not so sticky. Current researches on underwater adhesive materials mainly focused on natural adhesive proteins secreted by mussels and barnacles, but holdfast adhesive polysaccharide is more characteristic, which possesses a greater potential of development [12,13]. Its ultra-high viscosity can be applied to the field of safety devices with stringent requirements for adhesion force, its high environmental adaptability can be applied to the environment (such as underwater or special interface) where adhesives cannot be used for bonding or the bonding effect is poor, its high biocompatibility can be applied to the medical field, especially the implantable medical devices, and its property of non-chemical pollution meets the requirements to the development of adhesive industry towards green environmental protection.
This review attempted to briefly summarize the recent progress in holdfast, to comb the findings related to its biosynthesis, and to provide assistance for further studies and artificial manufacture of holdfast. The components of holdfast are mainly N-acetylglucosamine. The mechanism of holdfast biosynthesis is very complex, including the synthesis, transport and secretion of polysaccharides, as well as various modifications. Many genes are involved in the biosynthesis of holdfast; therefore, its regulation mode and pathways are extremely complex. In addition, the production of holdfast is also regulated by cell cycle and stress. Defining the biosynthetic mechanism of holdfast will help to understand why it is ultra-viscous and develop new viscous materials based on its properties.
COMPOSITION AND PHYSICOCHEMICAL CHARACTERISTICS OF HOLDFAST
The composition and chemical structure of holdfast are not yet completely expounded. A research provides a method to extract the holdfast from C. crescentus cultures and analyzes the monosaccharide components of the adhesive matrix, which includes Glucose, 3-O- methylglucose, mannose, N-acetylglucosamine, and xylose [14]. The treatment of holdfast with multiple enzymes reveals that the proteolytic enzymes can’t change its integrity and viscosity, while chitinase and lysozyme can destroy the viscosity of holdfast. It is known that the common substrates of chitinases and lysozymes include oligomers of N- acetylglucosamine (GlcNAc). After incubation with holdfast using a variety of fluorescein-conjugated lectins, respectively, only wheat germ agglutinin (WGA) can bind to holdfast.
The specific binding substrate of WGA is also GlcNAc, thus demonstrating that the main component of holdfast is not a protein but a polymer of polysaccharide with N-acetylglucosamine as the basic unit [15]. Meanwhile, when testing holdfast viscosity, the elastic strength of holdfast reduced to 10% of its original value after lysozyme treatment, demonstrating that GlcNAc is the main component of holdfast that produces viscosity [13]. Since holdfast retains a portion of its viscosity after lysozyme treatment, but the low viscosity of PGA is observed by comparison with the GlcNAc polymer poly-β-1, 6-N- acetylglucosamine (PGA), demonstrating that there may be other components affecting the high viscosity of holdfast [16]. In order to further clarify the chemical composition of holdfast, some researchers have used atomic force microscopy (AFM) to perform nanoindentation measurements on holdfast treated with different enzymes. It is found that the holdfast viscosity strength decreases about one-third after DNase I and lysozyme treatment at the same time, by referring to the viscosity change of holdfast after treatment,
indicating that DNA and GlcNAc may play an important role in its viscosity. However, the holdfast core stiffness is significantly decreased after treatment with proteinase K, demonstrating the presence of polypeptides in holdfast and their effect on the viscous strength [17]. In another study, researchers employed spectroscopic techniques to probe holdfast. Results indicate the similarity of the holdfast to peptidoglycan from other bacterial species; surface-sensitive sum frequency generation show that aromatic and hydroxyl groups related to this protein content at the adhesive interface could be playing a crucial role in adhesion [18]. In order to determine the size of holdfast, atomic force microscopy (AFM) and fluorescence microscopy are used to determine the final state of solidification after secretion of holdfast, which is disk-shaped, with a diameter of about 400 nm and a thickness of about 40 ~ 50 nm [19,20]. The holdfast polysaccharide has some gel-like properties, and the determination of the size of its two states of dryness and wetness by AFM has revealed that the air-dried holdfast is one-third of its original thickness [21]. Due to the less secretion of holdfast and its difficulty in purification, the available experimental techniques are still insufficient to fully resolve its chemical components, and even less to know about its precise structural features.
The means to determine the viscous strength of trace substances include techniques such as laser tweezers, atomic force microscopy, flow chamber shear pressure and micropipetting [22-25]. Because of the low molecular weight and high viscous strength of holdfast, all the means above can’t measure the viscosity of holdfast directly. Tsang PH et al. designed an AFM-based holdfast viscosity test protocol, which first allowed Caulobacter crescentus to adhere to a micropipette tip, and then uses a micromanipulator to pull the bacteria vertically to separate the holdfast from stalk or micropipette tip. The magnitude of tensile force was calculated by the deformation distance of the micropipette tip at the time of separation, and the maximum adhesion force of holdfast was up to 68N/mm2 (Figure 1). Their results showed that the location of the fracture was at the interface between holdfast and the bacteria, so it was presumed that its actual adhesive force might be higher than t hat of the result [26].
Figure 1: A single C. crescentus cell is attached to a thin flexible pipette. The micromanipulation holding suction pipette pulls the cell until detached. The force of adhesion can be calculated from the amount of bending required to break the cell–pipette contact.
BIOSYNTHESIS OF HOLDFAST
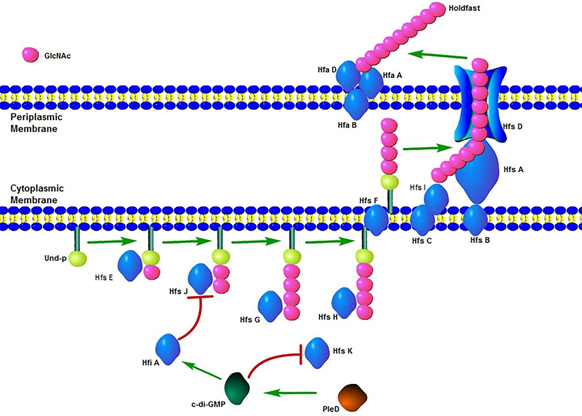
Through the combinatorial analysis of genes involved in the synthesis of holdfast, a biosynthetic pathway of holdfast can be roughly established. As a polysaccharide, the synthesis of holdfast starts with the transfer of glucose-1-phosphate from an uracil diphosphate (UDP) nuclear carrier to an undecaprenyl phosphate (Und-P) lipid carrier by the glycosyltransferase HfsE (holdfast synthesis E) to form monosaccharide glycolipids [27,28]. Then the glycosyltransferase HfsJ can synthesize monosaccharide lipids into disaccharide lipids, and the glycosyltransferase HfsG, which continues the work of HfsJ, further transfers N-acetylglucosamine to disaccharide lipids and further increasing the length of oligosaccharides. The deacetylase HfsH, partially deacetylates oligosaccharides, and both the synthesis and modification of these oligosaccharides are completed on the intracellular membrane. Subsequently, oligosaccharides cross the intracellular membrane into the periplasm by the flippase HfsF [29-31]. Oligosaccharides entering the cell periplasm are polymerized to form polysaccharides by the polymerase complex HfsC/I. After the polymerization of the polysaccharide, it is required to transport the polysaccharide to the outer membrane of the cell via the transmembrane transport complex HfsDAB [32,33]. Finally, the holdfast polysaccharide will localize to the anchoring protein complex HfaABD and undergo surface adhesion after secretion into the extracellular space through the transport complex. Subsequent secretion and adhesion processes are regulated by genes such as cell cycle-related factors and signal transduction [34-40]. A diagram of the holdfast biosynthetic pathway is shown in figure 2.
Figure 2: Diagram of Holdfast Biosynthesis Pathway. The synthesis of holdfast starts with the transfer of glucose-1-phosphate by the glycosyltransferase HfsE. Then the length of oligosaccharide is increased by HfsJ and HfsG. Oligosaccharide is partially deacetylated by the esterase HfsH. Oligosaccharide is transferred to the periplasm by the flippase HfsF and polymerized by the polymerase HfsC/I. Via the transmembrane transport complex HfsDAB, the holdfast polysaccharide is exported and localized to the anchoring protein complex HfaABD. The synthesis of holdfast is inhibited by HfiA which is regulated by c- di-GMP. The c-di-GMP also effects the N-acetyltransferase-like enzyme HfsK. PleD, the regulator of c-di-GMP contributes to holdfast induction along with flagellar assembly.
At present, some progress has been made in the study of its synthesis mechanism at the gene level, and more is known about the genes involved in the synthetic pathway of holdfast through whole-genome sequencing and analysis of Caulobacter Crescentus [41]. Genes directly involved in the synthesis and secretion of holdfast are divided into two gene clusters, named as holdfast synthesis (hfs) and holdfast anchoring (hfa) gene clusters [42,43]. On the other hand, the subsequent gene found to inhibit holdfast synthesisis named holdfast inhibitor (hfi) gene [44]. Functional analysis of these genes and their encoded proteins will help to further define the components, chemical modifications, and their detailed synthetic pathways of holdfast.
HOLDFAST POLYSACCHARIDE SYNTHESIS RELATED GENES
First, the initial step of holdfast synthesis is the transfer of glucose-1-phosphate from an uracil diphosphate (UDP) nuclear carrier to an undecaprenyl phosphate (Und-P) lipid carrier molecule by glycosyltransferase (HfsE) to form monosaccharide glycolipids. As the initial enzyme in the synthesis of holdfast, HfsE is replaceable. Synthesis of holdfast is not affected in the hfsE knockout strain. By genome sequence analysis, two hfsE homologs are found. Gene pssZ and pssY can express glycosyltransferases in strains of ΔhfsE [45].
The key gene in the synthesis of oligosaccharides from monosaccharide lipids is hfsJ, which encodes a glycosyltransferase that synthesizes monosaccharide lipids into disaccharide lipids. It has been shown that strains with a deletion mutation in hfsJ are unable to form holdfast, illustrating the irreplaceability of this gene [46]. Further studies have revealed that the gene hfiA, which has an inhibitory effect on holdfast synthesis, can specifically suppress the expression of hfsJ. At the same time, hfiA is regulated by multiple developmental regulatory proteins during cell cycle progression. Thus, hfiA is the connection of the holdfast synthesis pathway to the cell cycle regulatory network in Caulobacter Crescentus [47,48]. The hfsG gene, which encodes a glycosyltransferase, is responsible for transferring N-acetylglucosamine onto disaccharide lipids and further increasing the sugar chain length of oligosaccharides, while deletion of the hfsG gene results in failure of holdfast synthesis, demonstrating its critical role in holdfast synthesis [49].
Holdfast Modification and Transport Related Genes
After the oligosaccharide chains reach a certain length, the deacetylase encoded by the hfsH gene can partially deacetylate oligosaccharides. Studies have shown that strains with hfsH knockout can secrete holdfast normally, but it has almost no adhesive function. At the same time, strains overexpressing the hfsH gene have higher adhesion efficiency than that of wild-type strains [50, 51]. Studies of polysaccharide deacetylation have been proved its effect on viscosity enhancement, although the degree of deacetylation of holdfast polysaccharides is positively correlated with their viscosity, the upper limit of deacetylation cannot be determined. For example, galactosamine galactan outside the fungal cell mediates adhesion and biofilm formation by deacetylation, acetylated chitosan material mixed with chitin nanofibers improves the bond strength. Chitosan with a relatively high degree of deacetylation has higher bond strength and tensile strength compared with other adhesives [52-54]. Both the synthesis and modification of the oligosaccharides described above are completed on the inner membrane in the cytoplasm, and the oligosaccharides subsequently modified by deacetylation cross the intracellular membrane into the periplasm via the flippase encoded by the hfsF gene [55]. Oligosaccharides entering the periplasm of cells undergo polymerization to form polysaccharide chains by the polysaccharide copolymerase protein complex encoded by the hfsC/I gene, which is commonly found in the copolymerization transport system of exopolysaccharides and capsular polysaccharides of Gram-negative bacteria [56,57]. After completing the polymerization of polysaccharide chains, it is required to transport polysaccharides to the outer membrane of the cell through the transmembrane transport complex encoded by the holdfast transport-related gene cluster hfsDAB, which is unique in Caulobacter Crescentus. All the deletion mutants of hfsA, hfsB and hfsD show defects in adhesion function [58,59]. The protein encoded by hfsA has sequence similarity with the polysaccharide transporter GumC from X.Campestris, which has been demonstrated that GumC is involved in the transport of xanthan from the cytoplasm to the cell exterior [60]. HfsB and HfsD have no significant similarities to available protein sequences.
Holdfast Anchoring Related Genes
Synthetic holdfast polysaccharides will localize to the anchoring protein complex HfaABD and go to surface adhesion after secretion into the extracellular space through the transport complex. The occurrence and elongation of the organelle stalk for secretion of holdfast is associated with the cell cycle factor Pod J. In the swarmer cell the Pod J protein is located at the flagellar terminal in a short sequence. When the swarmer cell switches to the stalk cell, Pod J begins to synthesize the full-length form and localizes to terminal of stalk. The full-length Pod J can promote the elongation of stalk. Eventually the Pod J protein is processed into a short sequence in the late stalk cell and relocates to the dividing swarmer cell flagellar terminal after cell division [61,62]. During stalk formation for holdfast secretion, HfaABD protein complex begins to be synthesized and localized to the front of stalk extension. Gene hfaA, hfaB and hfaD are mutated and deleted, respectively, showing that holdfast would be released into the surrounding medium and unable to form adhesion. The hfaB mutant has the strongest holdfast shedding phenotype. HfaB and HfaD are shown to be membrane proteins by protein sequence analysis, while HfaA may have the ability to anchor holdfast to stalk tips [63,64]. Further studies on HfaABD protein complex show that their localization is mediated by both Pod J and Hfs (D, A, B) proteins, suggesting that the processes of holdfast synthesis, secretion, and anchoring are performed by a series of proteins in cooperation with each other [65].
EFFECT OF CELL CYCLE AND STRESS ON HOLDFAST
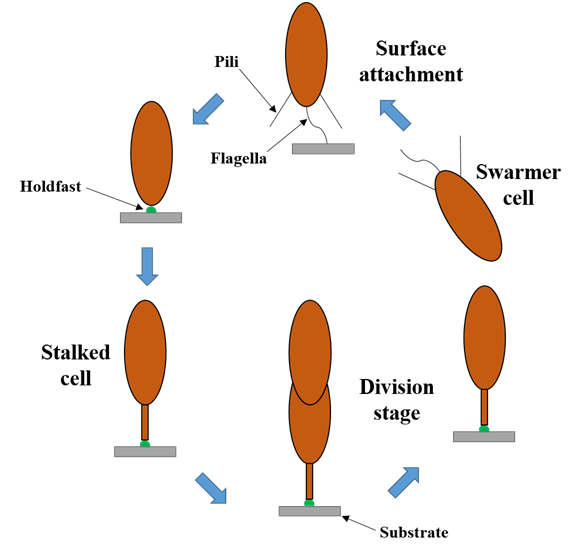
The process of secretion and adhesion of holdfast is closely related to cell cycle. There are two cell states in the growth process of Caulobacter Crescentus: swarmer cell and stalk cell. When the bacterium swims in water, it is in the swarmer cell state. When it contacts the surface of the medium to prepare for colonization, it elongates through the protrusions of the cell wall to form a structure called stalk, and permanently adheres to the surface of the medium after secretion of holdfast from the front end of stalk extension, and the cells in this state are called stalk cells [38]. After the stalk cell colonizes the surface of the medium and enters the next cell cycle to start dividing, the dividing cell leaves as a swarmer cell morphology. The transformation of these two cellular states is regulated by cell cycle- related signaling molecules and accompanied by the regulation of holdfast synthesis and secretion [39,40], the different states of Caulobacter Crescentus during the cell cycle are as follows (Figure 3). The synthesis and secretion of holdfast is not carried out during the whole cell cycle, and its initiation signal comes from the contact stimulation of the swarmer cell flagellum with the medium surface [66,67]. Studies on flagella have shown that in contrast to complete media, Caulobacter Crescentus does not stimulate holdfast synthesis due to its inability to synthesize flagella in defined media. Moreover, mutant strains lacking flagella adhere more efficiently to media surfaces than wild-type strains over time in defined media [68]. For this phenomenon, the current explanation is that the synthesis of c-di-GMP is downregulated due to decrease of PleD, a regulator of diguanylate cyclase, in the flagellar mutant, while transcription of the holdfast repressed gene hfiA is dependent on c-di-GMP, and eventually the synthesis of holdfast is initiated prematurely due to reduced expression of hfiA (Figure 4) [69]. Another research also discovers that PleD contributes to holdfast induction along with flagellar assembly as a developmental regulator, and proposes a model through which the flagellum integrates mechanical stimuli into the C. crescentus developmental program to coordinate adhesion [70]. During the study of the effect of c-di-GMP on holdfast synthesis, a new gene is identified named hfsK. The hfsK gene encodes an N-acetyltransferase-like enzyme which is also an effector of c-di-GMP. HfsK is localized in the cytoplasm by c-di- GMP mediation and inactivated at high concentrations of c-di-GMP, but the specific function of hfsK in the pathway of holdfast synthesis has not been clearly defined [71].
Figure 3: The cell cycle of Caulobacter crescentus. Surface contact stimulates the development of holdfast and stalk.
Figure 4: Stimulation contact of Flagellum and regulation of holdfast (HF) synthesis.Surface attachment results in a decrease of Pled and an increase of c-di-GMP, which reduces transcription of the holdfast synthesis inhibitor gene hfiA, thereby resulting in an increase in holdfast synthesis.
CONCLUSION AND PROSPECT
In conclusion, although the composition and precise structure of holdfast have not been fully elucidated, and its biosynthetic pathway has not been clear yet, the synthetic mechanism of holdfast in Caulobacter crescentus can be deeply studied at multiple levels with multi-disciplinary technologies in the future. Microbial engineering techniques can be used to solve the enrichment problem of holdfast, and obtaining a certain amount of this substance is the basis for conducting subsequent related studies. The synthetic pathway of holdfast can be regulated and simulated by means of genetic engineering as well as bio-manufacturing techniques, which can increase the yield of holdfast. New techniques such as structural biology and crystal analysis can be used to further understand the structural features of holdfast, mass spectrometry and nuclear magnetic resonance techniques can be applied in analyzing the complete chemical composition and structure of holdfast. Using genomics and bioinformatics techniques, the whole genome sequencing and functional analysis of Caulobacter crescentus can be performed to identify potential holdfast synthesis and modification-related genes, and further refine the understanding of its biosynthetic pathway. In the future, biomimetic adhesives will focus on the research and development of multi- material mixing substances. Adhesives inspired by mussel, barnacle, sandcastle worm and bacteria can combine the advantages of protein, polysaccharides and other adhesive components.With the rapid development of structural biology, material science and synthetic biology, the composition, structure and physicochemical characteristics of holdfast will be completely elucidated, and the clarification of its biosynthetic pathway and regulatory mechanism will lay the foundation for holdfast application and provide a new strategy for the development of novel biomimetic viscous materials.
REFERENCES
- Merker RI, Smit J. (1988). Characterization of the adhesive holdfast of marine and freshwater caulobacteria. Appl Environ Microbiol. 54(8):2078-85.
- Tsang PH, Li G, Brun YV, Freund LB, Tang JX. (2006). Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci U S A. 103(15):5764-8.
- Hauf M, Richter F, Schneider T, Faidt T, Martins BM, et al. (2017). Photoactivatable Mussel-based Underwater Adhesive Proteins by an Expanded Genetic Code. Chembiochem. 18(18):1819-1823.
- Lin Q, Gourdon D, Sun C, Holten-Andersen N, Anderson TH, et al. (2007). Adhesion mechanisms of the mussel foot proteins mfp-1 and mfp-3. Proc Natl Acad Sci U S A. 104 (10): 3782-6.
- Wei W, Yu J, Gebbie MA, Tan Y, Rodriguez NRM, et al. (2015). Bridging adhesion of mussel-inspired peptides: role of charge, chain length, and surface type. Langmuir. 31(3):1105-12.
- Nakano M, Kamino K. (2015). Amyloid-like conformation and interaction for the self-assembly in barnacle underwater cement. Biochemistry. 54(3):826-35.
- Liu X, Liang C, Zhang X, Li J, Huang J, et al. (2017). Amyloid fibril aggregation: An insight into the underwater adhesion of barnacle cement. Biochem Biophys Res Commun. 493(1):654-659.
- Liang C, Li Y, Liu Z, Wu W, Hu B. (2015). Protein Aggregation Formed by Recombinant cp19k Homology of Balanus albicostatus Combined with an 18 kDa N-Terminus Encoded by pET-32a (+) Plasmid Having Adhesion Strength Comparable to Severe Commercial Glues. PLoS One. 10(8):e0136493.
- Liang C, Ye Z, Xue B, Zeng L, Wu W, et al. (2018). Self-Assembled Nanofibers for Strong Underwater Adhesion: The Trick of Barnacles. ACS Appl Mater Interfaces. 10(30):25017-25025.
- Rocha M, Antas P, Castro LFC, Campos A, Vasconcelos V, et al. (2019). Comparative Analysis of the Adhesive Proteins of the Adult Stalked Goose Barnacle Pollicipes pollicipes (Cirripedia: Pedunculata). Mar Biotechnol (NY). 21(1):38-51.
- Brennan MJ, Kilbride BF, Wilker JJ, Liu JC. (2017). A bioinspired elastin-based protein for a cytocompatible underwater adhesive. Biomaterials. 124:116-125.
- Poindexter JS. (1964). Biological properties and classification of the Caulobacter group. Bacteriol Rev. 28:231-295.
- Li G, Smith CS, Brun YV, Tang JX. (2005). The elastic properties of Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J Bacteriol. 187(1):257-65.
- Hershey DM, Porfírio S, Black I, Jaehrig B, Heiss C, et al. (2019). Composition of the Holdfast Polysaccharide from Caulobacter crescentus. J Bacteriol. 201(17):e00276-19.
- Berne C, Ma X, Licata NA, Neves BRA, Setayeshgar S, et al. (2013). Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J Phys Chem B. 117(36):10492-503.
- Brown PJ, Hardy GG, Trimble MJ, Brun YV, et al. (2009). Complex regulatory pathways coordination cell-cycle progressionand development in Caulobacter crescentus. Adv Microb Physiol. 54:1-101.
- Hernando-Pérez M, Setayeshgar S, Hou Y, Temam R, Brun YV, et al. (2018). Layered Structure and Complex Mechanochemistry Underlie Strength and Versatility in a Bacterial Adhesive. M Bio. 9(1).
- Nyarko A, Singla S, Barton HA, Dhinojwala A. (2020). Spectroscopic Identification of Peptide Chemistry in the Caulobacter crescentus Holdfast. Biochemistry. 59(37):3508-3516.
- Li G, Brun YV, Tang JX. (2013). Holdfast spreading and thickening during Caulobacter crescentus attachment to surfaces. BMC Microbiol. 13:139.
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, et al. (2001). Complete genome sequence of Caulobacter Crescentus. Proc Natl Acad Sci USA. 98(7):4136-41.
- Toh E, Kurtz HD Jr, Brun YV. (2008). Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway replicates significant reduction in the initiating glycosyltransferase and polymerase steps. J Bacteriol. 190(21):7219-31.
- Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, et al. (2012). Functional characterization of UDP-glucose: undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter Crescentus. J Bacteriol. 194(10):2646-57.
- Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, et al. (2014). A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 10(1):e1004101.
- Eaton DS, Crosson S, Fiebig A. (2016). Proper Control of Caulobacter crescentus Cell Surface Adhesion Requirements the General Protein Chaperone DnaK. J Bacteriol. 198(19):2631-42.
- Wan Z, Brown PJ, Elliott EN, Brun YV. (2013). The adhesive and cooperative properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol. 88(3):486-500.
- Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. (2003). Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol. 185(4):1432-42.
- Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. (2009). Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 73(1):155-77.
- Javens J, Wan Z, Hardy GG, Brun YV. (2013). Bypassing the need for subcellular localization of a polysaccharide export-anchor complex by overexpression its protein subunits. Mol Microbiol. 89(2):350-71.
- Cole JL, Hardy GG, Bodenmiller D, Toh E, Hinz A, et al. (2003). The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol Microbiol. 49(6):1671-83.
- Sprecher KS, Hug I, Nesper J, Pothoff E, Mahi MA, et al. (2017). Cohesive properties of the Caulobacter crescentus holdfast adhesin are regulated by a novel c-di-GMP effector protein. MBio. 8(2).
- Hershey DM, Fiebig A, Crosson S. (2019). A Genome-Wide Analysis of Adhesion in Caulobacter crescentus Identifiers New Regulatory and Biosynthetic Components for Holder Fast Assembly. MBio. 10(1).
- Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, et al. (2012). Surface contact stimuli the just-in-time depletion of bacterial adhesins. Mol Microbiol. 83(1):41-51.
- Bodenmiller D, Toh E, Brun YV. (2004). Development of Surface Adhesion in Caulobacter crescentus. J Bacteriol. 186(5):1438-47.
- Hinz AJ, Larson DE, Smith CS, Brun YV. (2003). The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 47(4):929-41.
- Curtis PD, Quardokus EM, Lawler ML, Guo X, Klein D, et al. (2012). The scaffolding and signaling functions of a localization factor impact polar development. Mol Microbiol. 84(4):712-35.
- Hardy GG, Toh E, Berne C, Brun YV. (2018). Mutations in sugar-nucleotide synthesis genes restore holdfast polysaccharide anchoring to Caulobacter crescentus holdfast anchor mutants. J Bacteriol. 200(3).
- Hardy GG, Allen RC, Toh E, Brown PJB, Cole-Tobian JL, et al. (2010). A localized multimeric anchor attached to the Caulobacter holdfast to the cell pole. Mol Microbiol. 76(2):409-27.
- Berne C, Ellison C. K, Agarwal R, Severin GB, Fiebig A, et al. (2018). Feedback regulation of Caulobacter crescentus holdfast synthesis by flagellum assembly via the holdfast inhibitor HFIA. Mol Microbiol. 110(2):219-238.
- Hershey DM, Fiebig A, Crosson S. A Genome-Wide Analysis of Adhesion in Caulobacter crescentus Identifiers New Regulatory and Biosynthetic Components for Holder Fast Assembly.MBio.2019 Feb 12;10 (1).
- Bharat TAM, Kureisaite-Ciziene D, Hardy GG, Yu EW, Devant JM, et al. (2017). Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat Microbiol. 2:17059.
- Martins-Pinheiro M, Oliveira AR, Valencia AO, Fernandez-Silva FS, Silva LG, et al. (2017). Molecular characterization of Caulobacter crescentus mutator strains. Gene. 626:251-257.
- Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, et al. (2017). Observation of pilus retraction stimuli bacterial surface sensing. Science. 358:535-538.
- Hug I, Deshpande S, Sprecher KS, Pfohl T, Jenal U. (2017). Second messenger-mediated tactile response by a bacterial root-motor. Science. 358:531-534.
- Kurtz HD, Jr. Smit J. (1992). Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell.J Bacteriol. 174:687-694.
- Umbreit TH, Pate JL. (1978). Characterization of the holdfast region of wild-type cells and holdfast mutants of Asticcacaulis biprosthecum. Arch Microbiol. 118:157-168.
- Ong CJ, Wong ML, Smit J. (1990). Attachment of the adhesive holdfast organism to the cellular stalk of Caulobacter crescentus. J Bacteriol. 172:1448-1456.
- Levi A, Jenal U. (2006). Holder fast formation in mobile swarmer cells options surface attachment during Caulobacter crescentus development. J Bacteriol. 188:5315-5318.
- Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, et al. (2018). Mutant phenotypes for this population of bacterial genes of unknown function. Nature. 557:503-509.
- Hoffman MD, Zucker LI, Brown PJB, Kysela DT, Brun YV, et al. (2015). Timescales and frequencies of reversed and irreversible adhesion events of single bacterial cells. Anal Chem. 87:12032-12039.
- Leclerc G, Wang SP, Ely B. (1998). A new class of Caulobacter crescentus flagellar genes. J Bacteriol. 180:5010-5019.
- Abel S, Bucher T, Nicollier M, Hug I, Kaever V, et al. (2013). Bi-modal distribution of the second messenger c-di-GMP controls cell fate and apheresis during the Caulobacter cell cycle. PLoS Genet. 9:e1003744-17.
- Hentchel KL, Ruiz LMR, Curtis PD, Fiebig A, Coleman ML, et al. (2009). Genome-scale fitness profile of Caulobacter crescentus growth in natural freshwater. ISME J. 13:523- 536.
- Fang HH, Chan KY, Xu LC. (2000). Quantification of bacterial adhesion forces using atomic force microscopy (AFM). J Microbiol Methods. 40(1):89-97.
- Sagvolden G, Giaever I, Pettersen EO, Feder J. (1999). Cell adhesion force microscopy. Proc Natl Acad Sci U S A. 96(2):471-6.
- Burmeister JS, Vrany JD, Reichert WM, Truskey GA. (1996). Effect of fibronectin amount and formation on the strength of endothelial cell adhesion to HEMA/EMA copies. J Biomed Mater Res. 30(1):13-22.
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. (1999). Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 126(23):5431-40.
- Danner EW, Kan Y, Hammer MU, Israelachvili JN, Waite JH. (2012). Adhesion of muscle foot protein Mefp-5 to mica: an underwater superglue. Biochemistry. 51(33):6511-8.
- Lu Q, Danner E, Waite JH, Israelachvili JN, Zeng H, et al. (2013). Adhesion of muscle foot proteins to different substrate surfaces. J R Soc Interface. 10(79):20120759.
- Wei Q, Achazi K, Liebe H, Schulz A, Michael PL, et al. (2014). Mussel-inspired dendritic polymers as universal multifunctional coatings. Angew Chem Int Ed Engl. 53(43):11650-5.
- Liang C, Li Y, Liu Z, Wu W, Hu B. (2015). Protein Aggregation Formed by Recombinant cp19k Homology of Balanus albicostatus Combined with an 18 kDa N-Terminus Encoded by pET-32a (+) Plasmid Having Adhesion Strength Comparable to Severe Commercial Glues. PLoS One. 10(8):e0136493.
- Kurniasih M, Purwati I, Cahyati T, Dewi RS. (2018). Carboxymethyl chitosan as an antifungal agent on gauze. Int J Biol Macromol. 119:166-171.
- Younes I, Rinaudo M. (2015). Chitin and chitosan preparation from marine sources.Structure properties and applications. Mar Drugs. 13(3):1133-74.
- Neuman MG, Nanau RM, Oruña-Sanchez L, Coto G. (2015). Hyaluronic acid and wound healing. J Pharm Pharm Sci. 18(1):53-60.
- Sudha PN, Rose MH. (2014). Biological effects of hyaluronic acid. Adv Food Nut Res. 72:137-176.
- Persson A, Tykesson E, Westergren-Thorsson G, Malmström A, Ellervik U, et al. (2016). Xyloside-primed chondroitin sulfate/Dermatan sulfate from breast carcinoma cells with a defined polysaccharide composition has cytotoxic effects in Vitro. J Biol Chem. 291(28):14871-82.
- Dhahri M, Rodriguez-Ruiz V, Aid-Launais R, Ollivier V, Pavon-Djavid G, et al. (2017). In vitro and in vivo hemocompatibility evaluation of a new dermatan sulfate-modified PET patch for vascular repair surgery. J Biomed Mater Res B Appl Biomater. 105(7):2001-2009.
- Bougatef H, Krichen F, Capitani F, Amor IB, Maccari F, et al. (2015). Chondroitin sulfate/dermatan sulfate from corb (Sciaena umbra) skin: purification, structural analysis and anticoagulant effect. Carbohydr Polym. 196:272-278.
- Lee MJ, Geller AM, Bamford NC. (2016). Deacetylation of Fungal Exopolysaccharide Mediates Adhesion and Biofilm Formation. MBio. 7(2):e00252-16.
- Azuma K, Nishihara M, Shimizu H, Itoh Y, Takashima O, et al. (2015). Biological adhesive based on carboxymethyl chitin derivatives and chitin nanofibers. Biomaterials. 42:20-9.
- Hershey DM, Fiebig A, Crosson S. (2021). Flagellar Perturbations Activate Adhesion through Two Distinct Pathways in Caulobacter crescentus. mBio. 12(1):e03266-20.
- Barton MJ, Morley JW, Mahns DA, Mawad D, Wuhrer R, et al. (2014). Tissue repair strength using chitosan additives with different physical-chemical characteristics. J Biophotonics. 7(11):948-55.
Copyright: Xu DG, et al. © (2021). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Citation: Xu DG, et al. (2021). Research Advances in Biosynthesis Mechanism of Ultra-High Adhesive Material - Holdfast. Material Sci. 3(1):09.
 Abstract
Abstract  PDF
PDF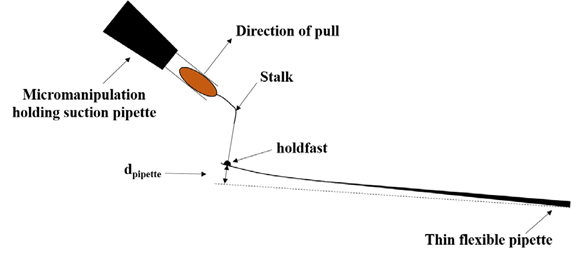
.png)